Germ Cell Development and Epigenetics
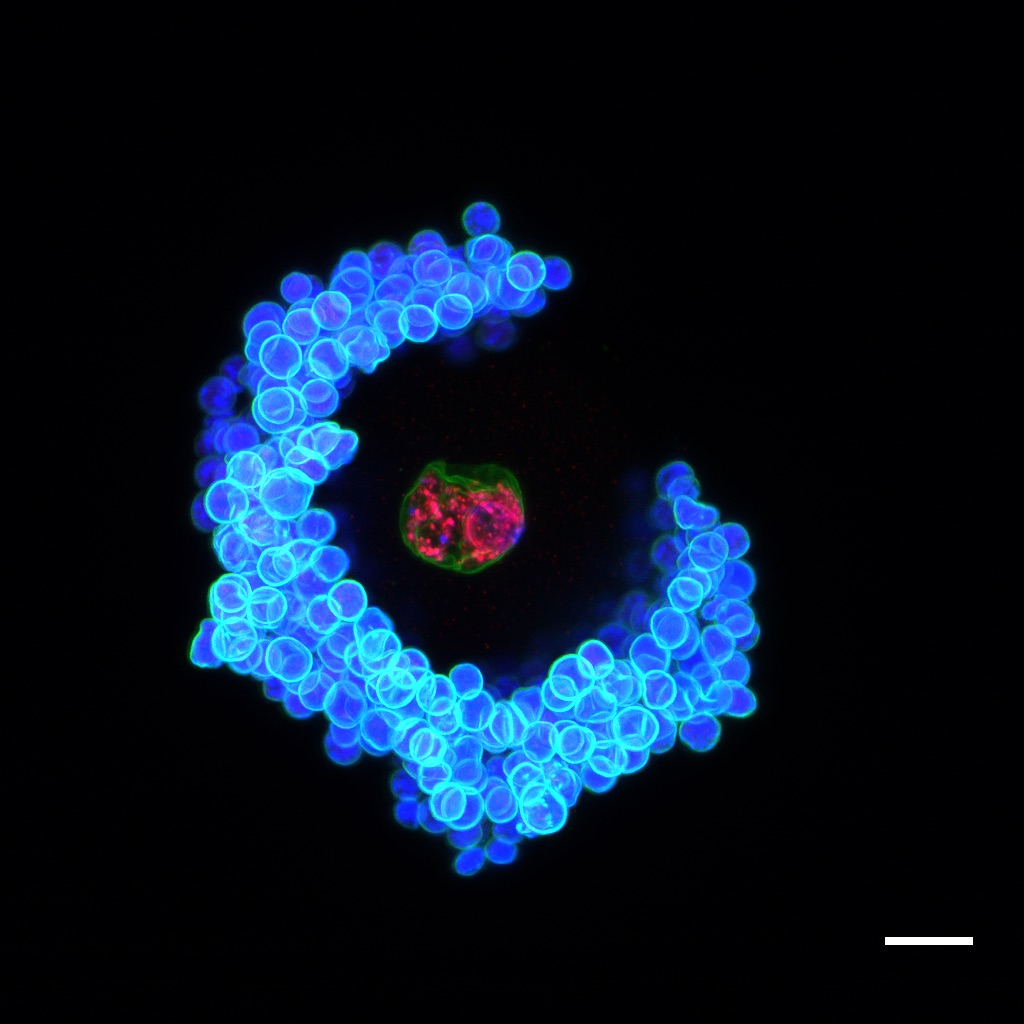
Oocyte (egg) surrounded by supporting cells
Overview
Germ cells are specialised cells found in the developing testes and ovaries that form sperm in males, or oocytes (eggs) in females. Sperm and oocytes transmit the parent’s genetic and epigenetic information to the offspring.
Epigenetic modifications to the chromatin (DNA plus the proteins that package it) provide a long-term “directory” or “memory” of which genes should be switched on or off in each cell, and thereby underpin cell identity and organ function. Conversely, disrupted epigenetic states occur in diseases including cancer, metabolic and behavioural disorders.
Importantly, epigenetic modifications are reversible in normal cells, allowing gene activity to be changed when necessary. This occurs most extensively in developing germ cells in which epigenetic information is re-set to equip the sperm and oocyte with the appropriate epigenetic information for directing embryonic and post-natal development in the offspring.
However, epigenetic programming is susceptible to alteration by environmental influences such as chemicals, diet and drugs. Significantly, altered epigenetic states can also be transmitted to the next generation and may affect health and development in the offspring. Such changes may contribute to the developmental origins of health and disease in a parent’s offspring.
The Germ Cell Development and Epigenetics group aims to improve understanding of epigenetics in the germ cells and the effects of epigenetic change on the offspring. Specifically, we use gene mutations and drugs to disrupt epigenetic modifier function in mouse germ cells to determine:
(i) the function of specific epigenetic modifiers in germ cell development, and
(ii) the ability of germ cells with altered epigenetic states to direct development in the parent’s offspring.
We also employ ex-vivo gonad culture and in-vivo mouse genetic models to examine the function of signalling pathways on epigenetics and gonad and germ cell development, providing insights into ovarian and testis cancers, and male and female infertility.
By exploring the establishment and function of epigenetic information in the germ line, our research will contribute to understanding human disease, including the impacts of novel drugs that target epigenetic processes.
Diseases we research
Research Group Head | Associate Professor Patrick Western
Oocytes (eggs) and sperm transmit the parents’ genetic and epigenetic information to the offspring. My research is focused on understanding the epigenetic processes underlying the establishment of the germ cell lineage and the impact of transmitted epigenetic information on subsequent generations.

News from the lab
Student opportunities

Publication highlights






